import uproot
print(f"uproot {uproot.__version__}")uproot 5.2.2Hans Dembinski, TU Dortmund
Build everything from basic components that fit together
Image credit: Toni Zaat on Unsplash
Python scientific stack
Other packages
Install everything with pip install LIBRARY
In terminal, create and enter virtual environment (you need Python-3.8 or later)
python3 -m venv .venv
source .venv/bin/activateInstall packages
python -m pip install --upgrade numba matplotlib uproot boost-histogram iminuit scipy particle numba_stats ipywidgetsDownload this notebook from indico and example.root, put latter into same folder
Run vscode on the current folder, then open the notebook inside the editor OR
Run
python -m pip install ipykernel
python -m ipykernel install --name "starterkit"
jupyter notebook lecture_1.ipynbThen select starterkit as kernel

name | typename | interpretation
---------------------+--------------------------+-------------------------------
trk_len | int32_t | AsDtype('>i4')
mc_trk_len | int32_t | AsDtype('>i4')
trk_imc | int32_t[] | AsJagged(AsDtype('>i4'))
trk_px | float[] | AsJagged(AsDtype('>f4'))
trk_py | float[] | AsJagged(AsDtype('>f4'))
trk_pz | float[] | AsJagged(AsDtype('>f4'))
mc_trk_px | float[] | AsJagged(AsDtype('>f4'))
mc_trk_py | float[] | AsJagged(AsDtype('>f4'))
mc_trk_pz | float[] | AsJagged(AsDtype('>f4'))
mc_trk_pid | int32_t[] | AsJagged(AsDtype('>i4'))Tree contains fake simulated LHCb events
Reconstructed tracks in simulation, branches with trk_*
Truth in simulation, branches with mc_trk_*
Some branches are 2D (e.g. trk_px)
Mathematical operators and slicing works on these arrays similar to Numpy
Many numpy functions work on awkward arrays
Exercise
np.sum and an array mask to count how many events have zero tracksawkward libraryExercise * Plot the branch trk_px with matplotlib.pyplot.hist * Can you figure out from the error message why it does not work? * Fix the issue with the function ak.flatten

┌───────────────────────────────────────────────────────────┐
[-inf, -2) 0 │ │
[ -2, -1.8) 1 │▏ │
[-1.8, -1.6) 3 │▎ │
[-1.6, -1.4) 17 │█▎ │
[-1.4, -1.2) 25 │█▊ │
[-1.2, -1) 78 │█████▌ │
[ -1, -0.8) 181 │████████████▉ │
[-0.8, -0.6) 308 │█████████████████████▊ │
[-0.6, -0.4) 453 │████████████████████████████████▏ │
[-0.4, -0.2) 647 │█████████████████████████████████████████████▉ │
[-0.2, 0) 819 │██████████████████████████████████████████████████████████ │
[ 0, 0.2) 782 │███████████████████████████████████████████████████████▍ │
[ 0.2, 0.4) 629 │████████████████████████████████████████████▌ │
[ 0.4, 0.6) 480 │██████████████████████████████████ │
[ 0.6, 0.8) 323 │██████████████████████▉ │
[ 0.8, 1) 159 │███████████▎ │
[ 1, 1.2) 77 │█████▌ │
[ 1.2, 1.4) 37 │██▋ │
[ 1.4, 1.6) 7 │▌ │
[ 1.6, 1.8) 3 │▎ │
[ 1.8, 2) 3 │▎ │
[ 2, inf) 0 │ │
└───────────────────────────────────────────────────────────┘locrebinTypical analysis work flow (often automated with Snakemake)
Many specialized fitting tools for individual purposes, e.g.: scipy.optimize.curve_fit
Generic libraries
Fitting (“estimation” in statistics)
Dataset: samples \(\{ \vec x_i \}\)
Model: Probability density or probability mass function \(f(\vec x; \vec p)\) which depends on unknown parameters \(\vec p\)
Conjecture: maximum-likelihood estimate (MLE) \(\hat{\vec p} = \text{argmax}_{\vec p} L(\vec p)\) is optimal, with \(L(\vec p) = \prod_i f(\vec x_i; \vec p)\)
Limiting case of maximum-likelihood fit: least-squares fit aka chi-square fit
'1.7.0'
# model pdf
def model1(x, z, mu, sigma, slope):
s = norm.pdf(x, mu, sigma)
b = truncexpon.pdf(x, 0.0, 1.0, 0.0, slope)
return (1 - z) * b + z * s
# we use NLL implementation provided by iminuit for convenience
from iminuit.cost import UnbinnedNLL
nll1 = UnbinnedNLL(x1, model1)
m = Minuit(nll1, z=0.5, mu=0.5, sigma=0.1, slope=1)| Migrad | |
|---|---|
| FCN = -4741 | Nfcn = 99 |
| EDM = 2.03e-05 (Goal: 0.0002) | |
| Valid Minimum | Below EDM threshold (goal x 10) |
| No parameters at limit | Below call limit |
| Hesse ok | Covariance accurate |
| Name | Value | Hesse Error | Minos Error- | Minos Error+ | Limit- | Limit+ | Fixed | |
|---|---|---|---|---|---|---|---|---|
| 0 | z | 0.489 | 0.009 | 0 | 1 | |||
| 1 | mu | 0.4983 | 0.0020 | 0 | 1 | |||
| 2 | sigma | 0.0977 | 0.0019 | 0 | ||||
| 3 | slope | 0.505 | 0.016 | 0 |
| z | mu | sigma | slope | |
|---|---|---|---|---|
| z | 8.48e-05 | -2e-6 (-0.102) | 9e-6 (0.535) | -0.06e-3 (-0.395) |
| mu | -2e-6 (-0.102) | 3.94e-06 | -0e-6 (-0.124) | -5e-6 (-0.153) |
| sigma | 9e-6 (0.535) | -0e-6 (-0.124) | 3.7e-06 | -9e-6 (-0.280) |
| slope | -0.06e-3 (-0.395) | -5e-6 (-0.153) | -9e-6 (-0.280) | 0.000268 |
Exercise
m.interactive() to check how fit reacts to parameter changesMinuit.mncontourExercise * Draw the likelihood profile contour of parameters z and sigma with Minuit.draw_mncontour * Compute matrix of likelihood profile contours with Minuit.draw_mnmatrix
numbaRooFit| Migrad | |
|---|---|
| FCN = 94.55 (χ²/ndof = 1.0) | Nfcn = 192 |
| EDM = 1.76e-07 (Goal: 0.0002) | |
| Valid Minimum | Below EDM threshold (goal x 10) |
| No parameters at limit | Below call limit |
| Hesse ok | Covariance accurate |
| Name | Value | Hesse Error | Minos Error- | Minos Error+ | Limit- | Limit+ | Fixed | |
|---|---|---|---|---|---|---|---|---|
| 0 | z | 501.0e-3 | 0.9e-3 | |||||
| 1 | mu | 500.3e-3 | 0.2e-3 | |||||
| 2 | sigma | 100.06e-3 | 0.20e-3 | |||||
| 3 | slope | 0.4982 | 0.0017 |
| z | mu | sigma | slope | |
|---|---|---|---|---|
| z | 8.91e-07 | -0.02e-6 (-0.103) | 0.10e-6 (0.557) | -0.7e-6 (-0.422) |
| mu | -0.02e-6 (-0.103) | 4.03e-08 | -0 (-0.125) | -0.05e-6 (-0.155) |
| sigma | 0.10e-6 (0.557) | -0 (-0.125) | 3.98e-08 | -0.10e-6 (-0.307) |
| slope | -0.7e-6 (-0.422) | -0.05e-6 (-0.155) | -0.10e-6 (-0.307) | 2.72e-06 |
ExtendedUnbinnedNLL or ExtendedBinnedNLLLeastSquaresTemplateiminuit.cost.Template
def make_data2(rng, nmc, truth, bins):
xe = np.linspace(0, 1, bins + 1)
b = np.diff(truncexpon.cdf(xe, 0, 1, 0, 1))
s = np.diff(norm.cdf(xe, 0.5, 0.05))
n = rng.poisson(b * truth[0]) + rng.poisson(s * truth[1])
t = np.array([rng.poisson(b * nmc), rng.poisson(s * nmc)])
return xe, n, t
rng = np.random.default_rng(1)
truth = 750, 250
xe, n, t = make_data2(rng, 200, truth, 15)
_, ax = plt.subplots(1, 2, figsize=(10, 4))
ax[0].stairs(n, xe, color="k", label="data")
ax[0].legend();
ax[1].stairs(t[0], xe, label="background")
ax[1].stairs(t[1], xe, label="signal")
ax[1].legend(title="templates");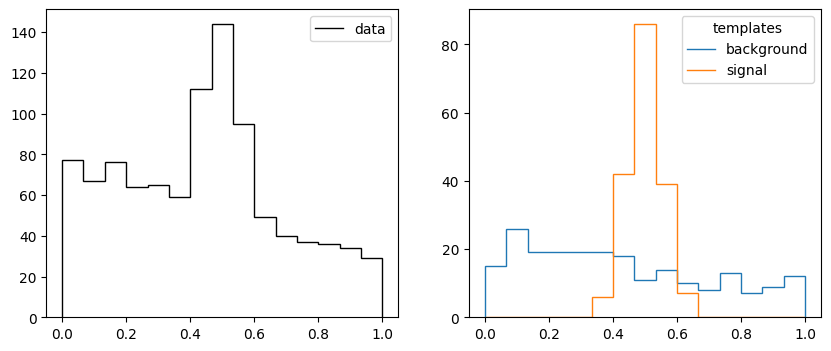
| Migrad | |
|---|---|
| FCN = 8.352 (χ²/ndof = 0.6) | Nfcn = 132 |
| EDM = 2.75e-07 (Goal: 0.0002) | |
| Valid Minimum | Below EDM threshold (goal x 10) |
| No parameters at limit | Below call limit |
| Hesse ok | Covariance accurate |
| Name | Value | Hesse Error | Minos Error- | Minos Error+ | Limit- | Limit+ | Fixed | |
|---|---|---|---|---|---|---|---|---|
| 0 | x0 | 770 | 70 | 0 | ||||
| 1 | x1 | 220 | 40 | 0 |
| x0 | x1 | |
|---|---|---|
| x0 | 4.3e+03 | -0.8e3 (-0.354) |
| x1 | -0.8e3 (-0.354) | 1.32e+03 |
Template you can also fit a mix of template and parametric modelExercise
make_data2 generate templates with 1 000 000 simulated pointsTemplate cost function