import uproot
print(f"uproot {uproot.__version__}")uproot 5.2.2Hans Dembinski, TU Dortmund
Build everything from basic components that fit together
Image credit: Toni Zaat on Unsplash
Python scientific stack
Other packages
Install everything with pip install LIBRARY
In terminal, create and enter virtual environment (you need Python-3.8 or later)
python3 -m venv .venv
source .venv/bin/activateInstall packages
python -m pip install --upgrade numba matplotlib uproot boost-histogram iminuit scipy particle numba_stats ipywidgets
name | typename | interpretation
---------------------+--------------------------+-------------------------------
trk_len | int32_t | AsDtype('>i4')
mc_trk_len | int32_t | AsDtype('>i4')
trk_imc | int32_t[] | AsJagged(AsDtype('>i4'))
trk_px | float[] | AsJagged(AsDtype('>f4'))
trk_py | float[] | AsJagged(AsDtype('>f4'))
trk_pz | float[] | AsJagged(AsDtype('>f4'))
mc_trk_px | float[] | AsJagged(AsDtype('>f4'))
mc_trk_py | float[] | AsJagged(AsDtype('>f4'))
mc_trk_pz | float[] | AsJagged(AsDtype('>f4'))
mc_trk_pid | int32_t[] | AsJagged(AsDtype('>i4'))Tree contains fake simulated LHCb events
Truth in simulation
Reconstructed simulation
Special
[6, 7, 2, 6, 7, 7, 6, 7, 4, 6, ..., 2, 4, 13, 3, 6, 3, 4, 6, 3] ------------------ type: 1000 * int32
Exercise: Explore some other arrays from event
0 6 [-0.979, 0.232, -0.464, 0.629, 0.0287, 0.156]
1 7 [-0.59, 0.102, -0.282, -0.585, 0.0525, -0.249, -0.0836]
2 2 [0.124, 0.309]
3 6 [-0.287, 0.0082, -0.671, -0.933, -0.011, 0.0323]
4 7 [-0.086, -0.101, 0.0523, 0.204, -0.0287, 0.115, 0.0954][False, False, False, False, False, False, False, False, False, False, ..., False, False, True, False, False, False, False, False, False] ----------------- type: 1000 * bool
Exercise
np.sum and array maskExercise * Try to plot trk_px * Can you figure out from the error message why it does not work? * Fix the issue with the function ak.flatten
%%timeit -n 1 -r 3
delta_px = []
for chunk in event.iterate(["trk_px", "trk_imc", "mc_trk_px"]):
for px, imc, mc_px in zip(chunk["trk_px"], chunk["trk_imc"], chunk["mc_trk_px"]):
# select only tracks with associated true particle
mask = imc >= 0
subset_px = px[mask]
subset_mc_px = mc_px[imc[mask]]
# conversion to numpy needed
delta_px = np.append(delta_px, subset_px - subset_mc_px) 963 ms ± 35.4 ms per loop (mean ± std. dev. of 3 runs, 1 loop each)# awkward arrays can be used in Numba
@nb.njit
def calc(chunk):
r = []
# loop over events
for px, imc, mc_px in zip(chunk["trk_px"], chunk["trk_imc"], chunk["mc_trk_px"]):
# loop over reconstructed tracks per event
for px_i, imc_i in zip(px, imc):
if imc_i < 0:
continue
mc_px_i = mc_px[imc_i]
r.append(px_i - mc_px_i)
return r4.17 ms ± 1.17 ms per loop (mean ± std. dev. of 3 runs, 1 loop each)delta_px could still become very large in memory
┌───────────────────────────────────────────────────────────┐
[-inf, -2) 0 │ │
[ -2, -1.8) 1 │▏ │
[-1.8, -1.6) 3 │▎ │
[-1.6, -1.4) 17 │█▎ │
[-1.4, -1.2) 25 │█▊ │
[-1.2, -1) 78 │█████▌ │
[ -1, -0.8) 181 │████████████▉ │
[-0.8, -0.6) 308 │█████████████████████▊ │
[-0.6, -0.4) 453 │████████████████████████████████▏ │
[-0.4, -0.2) 647 │█████████████████████████████████████████████▉ │
[-0.2, 0) 819 │██████████████████████████████████████████████████████████ │
[ 0, 0.2) 782 │███████████████████████████████████████████████████████▍ │
[ 0.2, 0.4) 629 │████████████████████████████████████████████▌ │
[ 0.4, 0.6) 480 │██████████████████████████████████ │
[ 0.6, 0.8) 323 │██████████████████████▉ │
[ 0.8, 1) 159 │███████████▎ │
[ 1, 1.2) 77 │█████▌ │
[ 1.2, 1.4) 37 │██▋ │
[ 1.4, 1.6) 7 │▌ │
[ 1.6, 1.8) 3 │▎ │
[ 1.8, 2) 3 │▎ │
[ 2, inf) 0 │ │
└───────────────────────────────────────────────────────────┘Exercise * Use iterative loop to fill h_mc_px and h_delta_px, use calc from before * Plot all three histograms overlaid into one figure with plt.stairs
# solution
for chunk in event.iterate(["trk_px", "trk_imc", "mc_trk_px"]):
h_px.fill(ak.flatten(chunk["trk_px"]))
h_mc_px.fill(ak.flatten(chunk["trk_px"]))
h_delta_px.fill(calc(chunk))
for h, label in ((h_px, "px"), (h_mc_px, "mc_px"), (h_delta_px, "delta_px")):
plt.stairs(h.values(), h.axes[0].edges, label=label)
plt.legend();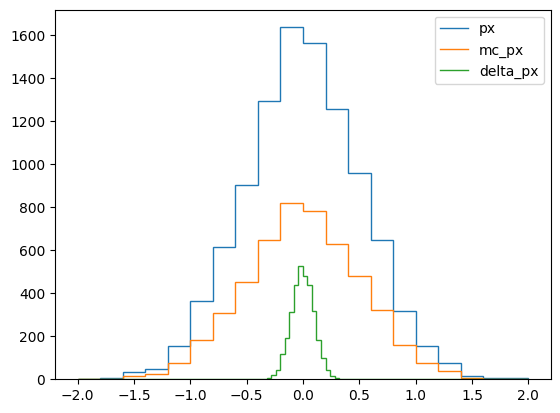
locrebinbranches = ["trk_px", "trk_imc", "mc_trk_px", "mc_trk_pid"]
for data in event.iterate(branches):
for px, imc, mc_px, mc_pid in zip(*(data[b] for b in branches)):
# select only tracks with associated true particle
mask = imc >= 0
px = px[mask]
associated = imc[mask]
mc_px = mc_px[associated]
mc_pid = mc_pid[associated]
p_2d.fill(mc_px, mc_pid, sample=px)
p_2dHistogram(
Regular(50, -2, 2),
IntCategory([-321, 211, 2212, -2212, -211, 321], growth=True),
storage=Mean()) # Sum: Mean(count=3240, value=0.000433322, variance=0.263444)particle library to look-up properties of particles based on their particle IDpids = p_2d.axes[1]
for i, pid in enumerate(pids):
part = particle.Particle.from_pdgid(pid)
x = p_2d.axes[0].centers
y = p_2d.values()[:, i]
ye = p_2d.variances()[:, i] ** 0.5
plt.errorbar(x, y, ye, fmt="o", label=f"${part.latex_name}$ pid={int(part.pdgid)}")
plt.xlabel("mc_px")
plt.ylabel("px")
plt.legend();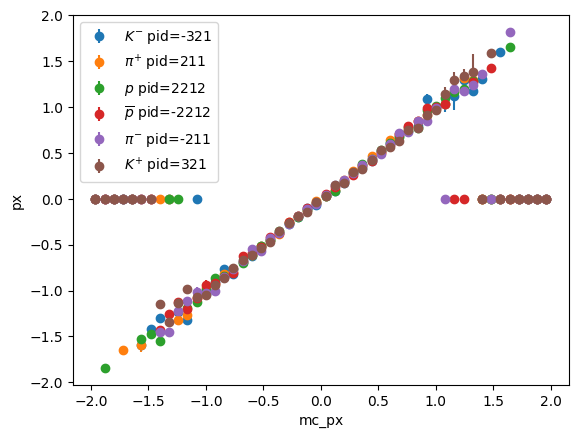
Exercise * Only show bins with sufficient entries, use p_2d.counts()
# solution
pids = p_2d.axes[1]
for i, pid in enumerate(pids):
part = particle.Particle.from_pdgid(pid)
x = p_2d.axes[0].centers
y = p_2d.values()[:, i]
ye = p_2d.variances()[:, i] ** 0.5
ma = (p_2d.counts() > 5)[:, i]
plt.errorbar(x[ma], y[ma], ye[ma], fmt="o",
label=f"${part.latex_name}$ pid={int(part.pdgid)}")
plt.xlabel("mc_px")
plt.ylabel("px")
plt.legend();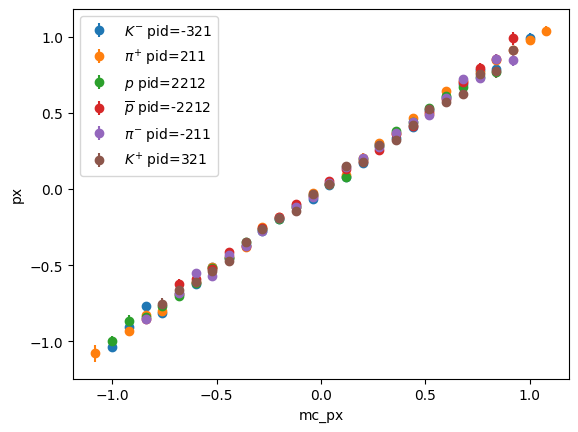
Typical analysis work flow (often automated with Snakemake)
Many specialized fitting tools for individual purposes, e.g.: scipy.optimize.curve_fit
Generic libraries
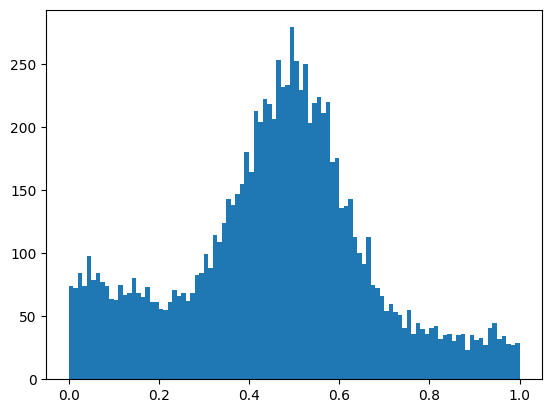
Fitting (“estimation” in statistics)
Dataset: samples \(\{ \vec x_i \}\)
Model: Probability density or probability mass function \(f(\vec x; \vec p)\) which depends on unknown parameters \(\vec p\)
Conjecture: maximum-likelihood estimate (MLE) \(\hat{\vec p} = \text{argmax}_{\vec p} L(\vec p)\) is optimal, with \(L(\vec p) = \prod_i f(\vec x_i; \vec p)\)
Limiting case of maximum-likelihood fit: least-squares fit aka chi-square fit
# fast implementations of statistical distributions, API similar to scipy.stats
from numba_stats import norm, truncexpon
# model pdf
def model1(x, z, mu, sigma, slope):
s = norm.pdf(x, mu, sigma)
b = truncexpon.pdf(x, 0.0, 1.0, 0.0, slope)
return (1 - z) * b + z * s
# negative log-likelihood
def nll1(z, mu, sigma, slope):
logL = np.log(model1(x1, z, mu, sigma, slope))
return -np.sum(logL)| Migrad | |
|---|---|
| FCN = -1790 | Nfcn = 209 |
| EDM = 762 (Goal: 0.0001) | |
| INVALID Minimum | ABOVE EDM threshold (goal x 10) |
| No parameters at limit | Below call limit |
| Hesse ok | Covariance accurate |
| Name | Value | Hesse Error | Minos Error- | Minos Error+ | Limit- | Limit+ | Fixed | |
|---|---|---|---|---|---|---|---|---|
| 0 | z | 0.500 | 0.012 | |||||
| 1 | mu | 0.500 | 0.003 | |||||
| 2 | sigma | 0.155 | 0.005 | |||||
| 3 | slope | 1.00 | 0.07 |
| z | mu | sigma | slope | |
|---|---|---|---|---|
| z | 0.000149 | -1e-6 (-0.037) | 0.033e-3 (0.508) | -0.25e-3 (-0.304) |
| mu | -1e-6 (-0.037) | 9.19e-06 | -0e-6 (-0.025) | -68e-6 (-0.335) |
| sigma | 0.033e-3 (0.508) | -0e-6 (-0.025) | 2.91e-05 | -0.053e-3 (-0.147) |
| slope | -0.25e-3 (-0.304) | -68e-6 (-0.335) | -0.053e-3 (-0.147) | 0.00442 |
W FCN result is NaN for [ 0.406325 0.495731 -0.0437361 1.13819 ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W VariableMetricBuilder No improvement in line search
W VariableMetricBuilder Iterations finish without convergence; Edm 761.614 Requested 0.0001
W VariableMetricBuilder No convergence; Edm 761.614 is above tolerance 0.001
W FCN result is NaN for [ 0.406325 0.495731 -0.0437361 1.13819 ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W VariableMetricBuilder No improvement in line search
W VariableMetricBuilder Iterations finish without convergence; Edm 761.614 Requested 0.0001
W VariableMetricBuilder No convergence; Edm 761.614 is above tolerance 0.001
W FCN result is NaN for [ 0.406325 0.495731 -0.0437361 1.13819 ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W VariableMetricBuilder No improvement in line search
W VariableMetricBuilder Iterations finish without convergence; Edm 761.614 Requested 0.0001
W VariableMetricBuilder No convergence; Edm 761.614 is above tolerance 0.001
W FCN result is NaN for [ 0.406325 0.495731 -0.0437361 1.13819 ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W VariableMetricBuilder No improvement in line search
W VariableMetricBuilder Iterations finish without convergence; Edm 761.614 Requested 0.0001
W VariableMetricBuilder No convergence; Edm 761.614 is above tolerance 0.001
W FCN result is NaN for [ 0.406325 0.495731 -0.0437361 1.13819 ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W FCN result is NaN for [ nan nan nan nan ]
W VariableMetricBuilder No improvement in line search
W VariableMetricBuilder Iterations finish without convergence; Edm 761.614 Requested 0.0001
W VariableMetricBuilder No convergence; Edm 761.614 is above tolerance 0.001Exercise
m.limits["parameter"] = (lower, upper) before m.migrad()m.limits["a", "b"] = (0, 1)| Migrad | |
|---|---|
| FCN = -2095 | Nfcn = 481 |
| EDM = 1.78e-06 (Goal: 0.0001) | |
| Valid Minimum | Below EDM threshold (goal x 10) |
| No parameters at limit | Below call limit |
| Hesse ok | Covariance accurate |
| Name | Value | Hesse Error | Minos Error- | Minos Error+ | Limit- | Limit+ | Fixed | |
|---|---|---|---|---|---|---|---|---|
| 0 | z | 0.498 | 0.009 | 0 | 1 | |||
| 1 | mu | 0.4968 | 0.0020 | 0 | 1 | |||
| 2 | sigma | 0.0994 | 0.0020 | 0 | ||||
| 3 | slope | 1.03 | 0.06 | 0 |
| z | mu | sigma | slope | |
|---|---|---|---|---|
| z | 8.94e-05 | -1e-6 (-0.063) | 10e-6 (0.547) | -0.12e-3 (-0.213) |
| mu | -1e-6 (-0.063) | 4.21e-06 | -0e-6 (-0.061) | -27e-6 (-0.225) |
| sigma | 10e-6 (0.547) | -0e-6 (-0.061) | 3.94e-06 | -18e-6 (-0.156) |
| slope | -0.12e-3 (-0.213) | -27e-6 (-0.225) | -18e-6 (-0.156) | 0.00345 |
| Migrad | |
|---|---|
| FCN = -2095 | Nfcn = 692 |
| EDM = 1.78e-06 (Goal: 0.0001) | |
| Valid Minimum | Below EDM threshold (goal x 10) |
| No parameters at limit | Below call limit |
| Hesse ok | Covariance accurate |
| Name | Value | Hesse Error | Minos Error- | Minos Error+ | Limit- | Limit+ | Fixed | |
|---|---|---|---|---|---|---|---|---|
| 0 | z | 0.498 | 0.009 | -0.009 | 0.009 | 0 | 1 | |
| 1 | mu | 0.4968 | 0.0021 | -0.0020 | 0.0020 | 0 | 1 | |
| 2 | sigma | 0.0994 | 0.0020 | -0.0019 | 0.0020 | 0 | ||
| 3 | slope | 1.03 | 0.06 | -0.06 | 0.06 | 0 |
| z | mu | sigma | slope | |||||
|---|---|---|---|---|---|---|---|---|
| Error | -0.009 | 0.009 | -0.002 | 0.002 | -0.0019 | 0.0020 | -0.06 | 0.06 |
| Valid | True | True | True | True | True | True | True | True |
| At Limit | False | False | False | False | False | False | False | False |
| Max FCN | False | False | False | False | False | False | False | False |
| New Min | False | False | False | False | False | False | False | False |
| z | mu | sigma | slope | |
|---|---|---|---|---|
| z | 8.94e-05 | -1e-6 (-0.063) | 10e-6 (0.547) | -0.12e-3 (-0.213) |
| mu | -1e-6 (-0.063) | 4.21e-06 | -0e-6 (-0.061) | -27e-6 (-0.225) |
| sigma | 10e-6 (0.547) | -0e-6 (-0.061) | 3.94e-06 | -18e-6 (-0.156) |
| slope | -0.12e-3 (-0.213) | -27e-6 (-0.225) | -18e-6 (-0.156) | 0.00345 |
mncontour and draw them with draw_mncontourExercise
cldraw_mnmatrix| Migrad | |
|---|---|
| FCN = -4189 | Nfcn = 169 |
| EDM = 8.37e-05 (Goal: 0.0002) | time = 0.4 sec |
| Valid Minimum | Below EDM threshold (goal x 10) |
| No parameters at limit | Below call limit |
| Hesse ok | Covariance accurate |
| Name | Value | Hesse Error | Minos Error- | Minos Error+ | Limit- | Limit+ | Fixed | |
|---|---|---|---|---|---|---|---|---|
| 0 | z | 0.498 | 0.009 | 0 | 1 | |||
| 1 | mu | 0.4968 | 0.0020 | 0 | 1 | |||
| 2 | sigma | 0.0994 | 0.0020 | 0 | ||||
| 3 | slope | 1.03 | 0.06 | 0 |
| z | mu | sigma | slope | |
|---|---|---|---|---|
| z | 8.62e-05 | -1e-6 (-0.037) | 10e-6 (0.529) | -0.11e-3 (-0.211) |
| mu | -1e-6 (-0.037) | 4.17e-06 | -0e-6 (-0.039) | -28e-6 (-0.230) |
| sigma | 10e-6 (0.529) | -0e-6 (-0.039) | 3.83e-06 | -17e-6 (-0.152) |
| slope | -0.11e-3 (-0.211) | -28e-6 (-0.230) | -17e-6 (-0.152) | 0.00345 |
Exercise
m.interactive() to check how fit reacts to parameter changes| Migrad | |
|---|---|
| FCN = -4.216e+05 | Nfcn = 169 |
| EDM = 0.000135 (Goal: 0.0002) | time = 5.9 sec |
| Valid Minimum | Below EDM threshold (goal x 10) |
| No parameters at limit | Below call limit |
| Hesse ok | Covariance accurate |
| Name | Value | Hesse Error | Minos Error- | Minos Error+ | Limit- | Limit+ | Fixed | |
|---|---|---|---|---|---|---|---|---|
| 0 | z | 499.7e-3 | 0.9e-3 | 0 | 1 | |||
| 1 | mu | 499.98e-3 | 0.20e-3 | 0 | 1 | |||
| 2 | sigma | 99.59e-3 | 0.20e-3 | 0 | ||||
| 3 | slope | 0.997 | 0.006 | 0 |
| z | mu | sigma | slope | |
|---|---|---|---|---|
| z | 8.87e-07 | -0.01e-6 (-0.049) | 0.10e-6 (0.548) | -1.2e-6 (-0.229) |
| mu | -0.01e-6 (-0.049) | 4.18e-08 | -0 (-0.060) | -0.26e-6 (-0.226) |
| sigma | 0.10e-6 (0.548) | -0 (-0.060) | 4.09e-08 | -0.18e-6 (-0.164) |
| slope | -1.2e-6 (-0.229) | -0.26e-6 (-0.226) | -0.18e-6 (-0.164) | 3.06e-05 |
General insights * Time spend in minimizer is negligible * Bottleneck is evaluating NLL function * Bottleneck inside NLL function is usually evaluation of model * NLL computation is trivially parallelizable: log(pdf) computed independently for each data point * Numba allows us to exploit auto-vectorization (SIMD instructions) and parallel computing on multiple cors
@nb.njit(parallel=True, fastmath=True, error_model="numpy")
def nll2(z, mu, sigma, slope):
def model(x, z, mu, sigma, slope):
s = norm.pdf(x, mu, sigma)
b = truncexpon.pdf(x, 0.0, 1.0, 0.0, slope)
return (1 - z) * b + z * s
logL = np.log(model(x2, z, mu, sigma, slope))
return -np.sum(logL)
# compile function
nll2(0.5, 0.5, 0.1, 0.1);OMP: Info #276: omp_set_nested routine deprecated, please use omp_set_max_active_levels instead.| Migrad | |
|---|---|
| FCN = -2.108e+05 | Nfcn = 242 |
| EDM = 2.48e-06 (Goal: 0.0001) | time = 1.4 sec |
| Valid Minimum | Below EDM threshold (goal x 10) |
| No parameters at limit | Below call limit |
| Hesse ok | Covariance accurate |
| Name | Value | Hesse Error | Minos Error- | Minos Error+ | Limit- | Limit+ | Fixed | |
|---|---|---|---|---|---|---|---|---|
| 0 | z | 499.7e-3 | 0.9e-3 | 0 | 1 | |||
| 1 | mu | 499.98e-3 | 0.20e-3 | 0 | 1 | |||
| 2 | sigma | 99.59e-3 | 0.20e-3 | 0 | ||||
| 3 | slope | 0.997 | 0.006 | 0 |
| z | mu | sigma | slope | |
|---|---|---|---|---|
| z | 8.87e-07 | -0.01e-6 (-0.049) | 0.10e-6 (0.548) | -1.2e-6 (-0.229) |
| mu | -0.01e-6 (-0.049) | 4.18e-08 | -0 (-0.060) | -0.26e-6 (-0.226) |
| sigma | 0.10e-6 (0.548) | -0 (-0.060) | 4.09e-08 | -0.18e-6 (-0.164) |
| slope | -1.2e-6 (-0.229) | -0.26e-6 (-0.226) | -0.18e-6 (-0.164) | 3.06e-05 |
Often simpler: fit histograms
No bias from fitting histograms if done correctly, only loss in precision
Need model cdf instead of model pdf: \(F(x; \vec p) = \int_{-\infty}^x f(x'; \vec p) \text{d}x'\)
Recommended to use iminuit.cost.BinnedNLL, comes with many useful features
| Migrad | |
|---|---|
| FCN = 84.34 (χ²/ndof = 0.9) | Nfcn = 162 |
| EDM = 1.36e-07 (Goal: 0.0002) | time = 0.4 sec |
| Valid Minimum | Below EDM threshold (goal x 10) |
| No parameters at limit | Below call limit |
| Hesse ok | Covariance accurate |
| Name | Value | Hesse Error | Minos Error- | Minos Error+ | Limit- | Limit+ | Fixed | |
|---|---|---|---|---|---|---|---|---|
| 0 | z | 499.7e-3 | 0.9e-3 | |||||
| 1 | mu | 499.99e-3 | 0.20e-3 | |||||
| 2 | sigma | 99.58e-3 | 0.20e-3 | |||||
| 3 | slope | 0.997 | 0.006 |
| z | mu | sigma | slope | |
|---|---|---|---|---|
| z | 8.98e-07 | -0.01e-6 (-0.049) | 0.10e-6 (0.535) | -1.2e-6 (-0.236) |
| mu | -0.01e-6 (-0.049) | 4.22e-08 | -0 (-0.052) | -0.26e-6 (-0.228) |
| sigma | 0.10e-6 (0.535) | -0 (-0.052) | 4.15e-08 | -0.18e-6 (-0.161) |
| slope | -1.2e-6 (-0.236) | -0.26e-6 (-0.228) | -0.18e-6 (-0.161) | 3.09e-05 |
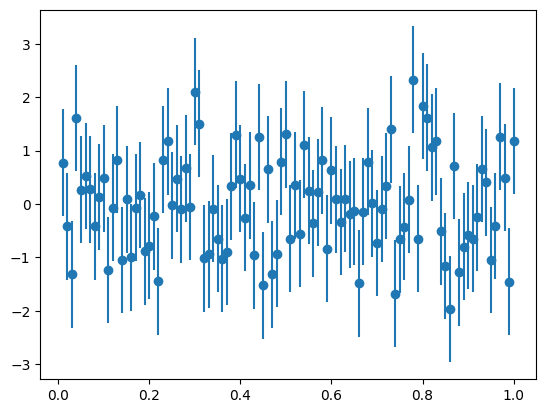
'chi2/ndof = 0.88 p-value = 0.80'Exercise
np.mean and np.std)ExtendedUnbinnedNLL or ExtendedBinnedNLLLeastSquaresTemplateiminuit.cost.Template
def make_data2(rng, nmc, truth, bins):
xe = np.linspace(0, 1, bins + 1)
b = np.diff(truncexpon.cdf(xe, 0, 1, 0, 1))
s = np.diff(norm.cdf(xe, 0.5, 0.05))
n = rng.poisson(b * truth[0]) + rng.poisson(s * truth[1])
t = np.array([rng.poisson(b * nmc), rng.poisson(s * nmc)])
return xe, n, t
rng = np.random.default_rng(1)
truth = 750, 250
xe, n, t = make_data2(rng, 200, truth, 15)
_, ax = plt.subplots(1, 2, figsize=(10, 4))
ax[0].stairs(n, xe, color="k", label="data")
ax[0].legend();
ax[1].stairs(t[0], xe, label="background")
ax[1].stairs(t[1], xe, label="signal")
ax[1].legend(title="templates");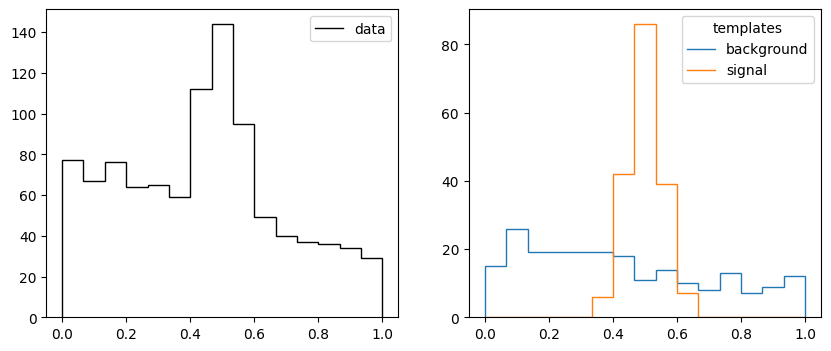
| Migrad | |
|---|---|
| FCN = 8.352 (χ²/ndof = 0.6) | Nfcn = 132 |
| EDM = 2.75e-07 (Goal: 0.0002) | time = 0.5 sec |
| Valid Minimum | Below EDM threshold (goal x 10) |
| No parameters at limit | Below call limit |
| Hesse ok | Covariance accurate |
| Name | Value | Hesse Error | Minos Error- | Minos Error+ | Limit- | Limit+ | Fixed | |
|---|---|---|---|---|---|---|---|---|
| 0 | x0 | 770 | 70 | 0 | ||||
| 1 | x1 | 220 | 40 | 0 |
| x0 | x1 | |
|---|---|---|
| x0 | 4.3e+03 | -0.8e3 (-0.354) |
| x1 | -0.8e3 (-0.354) | 1.32e+03 |
Exercise
make_data2 generate templates with 1 000 000 simulated pointsTemplate cost function| Migrad | |
|---|---|
| FCN = 4.329 (χ²/ndof = 0.3) | Nfcn = 110 |
| EDM = 2.66e-06 (Goal: 0.0002) | |
| Valid Minimum | Below EDM threshold (goal x 10) |
| No parameters at limit | Below call limit |
| Hesse ok | Covariance accurate |
| Name | Value | Hesse Error | Minos Error- | Minos Error+ | Limit- | Limit+ | Fixed | |
|---|---|---|---|---|---|---|---|---|
| 0 | x0 | 820 | 32 | 0 | ||||
| 1 | x1 | 201 | 20 | 0 |
| x0 | x1 | |
|---|---|---|
| x0 | 1.03e+03 | -0.2e3 (-0.326) |
| x1 | -0.2e3 (-0.326) | 415 |
Template you can also fit a mix of template and parametric model| Migrad | |
|---|---|
| FCN = 2.526 (χ²/ndof = 0.2) | Nfcn = 106 |
| EDM = 1.52e-06 (Goal: 0.0002) | |
| Valid Minimum | Below EDM threshold (goal x 10) |
| No parameters at limit | Below call limit |
| Hesse ok | Covariance accurate |
| Name | Value | Hesse Error | Minos Error- | Minos Error+ | Limit- | Limit+ | Fixed | |
|---|---|---|---|---|---|---|---|---|
| 0 | x0 | 819 | 34 | 0 | ||||
| 1 | x1_s | 202 | 23 | |||||
| 2 | x1_mu | 0.492 | 0.006 | 0 | 1 | |||
| 3 | x1_sigma | 0.049 | 0.007 | 0 |
| x0 | x1_s | x1_mu | x1_sigma | |
|---|---|---|---|---|
| x0 | 1.16e+03 | -0.3e3 (-0.428) | 8.13e-3 (0.038) | -79.31e-3 (-0.330) |
| x1_s | -0.3e3 (-0.428) | 541 | -8.17e-3 (-0.056) | 79.27e-3 (0.483) |
| x1_mu | 8.13e-3 (0.038) | -8.17e-3 (-0.056) | 3.99e-05 | -0 (-0.069) |
| x1_sigma | -79.31e-3 (-0.330) | 79.27e-3 (0.483) | -0 (-0.069) | 4.98e-05 |
from iminuit.cost import LeastSquares
from numba_stats import bernstein # Bernstein polynomials > Chebychev polynomials
x = [-0.37, -0.25, -0.15, -0.05, 0.047, 0.147, 0.243, 0.374]
y = [0.96, 0.90, 0.85, 0.81, 0.75, 0.66, 0.42, 0.14]
ey = [0.01, 0.01, 0.01, 0.02, 0.02, 0.03, 0.04, 0.05]
def model(x, p):
return bernstein.density(x, p, -0.4, 0.4)| Migrad | |
|---|---|
| FCN = 23.81 (χ²/ndof = 4.8) | Nfcn = 51 |
| EDM = 4.25e-21 (Goal: 0.0002) | |
| Valid Minimum | Below EDM threshold (goal x 10) |
| No parameters at limit | Below call limit |
| Hesse ok | Covariance accurate |
| Name | Value | Hesse Error | Minos Error- | Minos Error+ | Limit- | Limit+ | Fixed | |
|---|---|---|---|---|---|---|---|---|
| 0 | x0 | 0.950 | 0.011 | |||||
| 1 | x1 | 0.920 | 0.027 | |||||
| 2 | x2 | 0.21 | 0.04 |
| x0 | x1 | x2 | |
|---|---|---|---|
| x0 | 0.000127 | -0.21e-3 (-0.680) | 0.14e-3 (0.321) |
| x1 | -0.21e-3 (-0.680) | 0.000737 | -0.7e-3 (-0.666) |
| x2 | 0.14e-3 (0.321) | -0.7e-3 (-0.666) | 0.00147 |
Exercise
Fit with increasing number of parameters (increase the length of the starting parameters)
Try fitting this model
| Migrad | |
|---|---|
| FCN = 1.069 (χ²/ndof = 0.5) | Nfcn = 152 |
| EDM = 2.41e-15 (Goal: 0.0002) | |
| Valid Minimum | Below EDM threshold (goal x 10) |
| No parameters at limit | Below call limit |
| Hesse ok | Covariance accurate |
| Name | Value | Hesse Error | Minos Error- | Minos Error+ | Limit- | Limit+ | Fixed | |
|---|---|---|---|---|---|---|---|---|
| 0 | x0 | 0.956 | 0.026 | |||||
| 1 | x1 | 1.02 | 0.13 | |||||
| 2 | x2 | 0.47 | 0.27 | |||||
| 3 | x3 | 1.30 | 0.34 | |||||
| 4 | x4 | 0.29 | 0.25 | |||||
| 5 | x5 | 0.10 | 0.08 |
| x0 | x1 | x2 | x3 | x4 | x5 | |
|---|---|---|---|---|---|---|
| x0 | 0.000693 | -3.0e-3 (-0.885) | 5.7e-3 (0.800) | -6.2e-3 (-0.687) | 3.6e-3 (0.551) | -0.7e-3 (-0.307) |
| x1 | -3.0e-3 (-0.885) | 0.0161 | -0.033 (-0.961) | 0.037 (0.852) | -0.022 (-0.698) | 0.004 (0.394) |
| x2 | 5.7e-3 (0.800) | -0.033 (-0.961) | 0.0721 | -0.09 (-0.953) | 0.05 (0.822) | -0.010 (-0.480) |
| x3 | -6.2e-3 (-0.687) | 0.037 (0.852) | -0.09 (-0.953) | 0.116 | -0.08 (-0.938) | 0.016 (0.583) |
| x4 | 3.6e-3 (0.551) | -0.022 (-0.698) | 0.05 (0.822) | -0.08 (-0.938) | 0.062 | -0.014 (-0.721) |
| x5 | -0.7e-3 (-0.307) | 0.004 (0.394) | -0.010 (-0.480) | 0.016 (0.583) | -0.014 (-0.721) | 0.00649 |