from matplotlib import pyplot as plt
import numpy as np
mu = 1
sigma = 1
N = 10
K = 10000
var_1, var_2 = [], []
for k in range(K):
rng = np.random.default_rng(seed=k)
x_k = rng.normal(mu, sigma, size=N)
v1 = np.var(x_k)
v2 = np.mean((x_k - mu) ** 2)
var_1.append(v1)
var_2.append(v2)Hans Dembinski, TU Dortmund
- Uncertainties of estimators
- Bootstrapping of uncertainties
- Error propagation
- Estimation of systematic uncertainties
- Resources for further learning
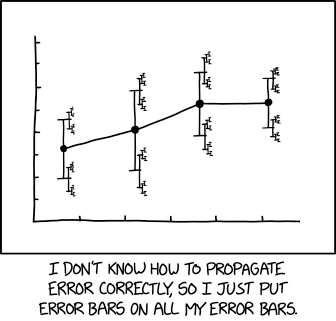
What is an uncertainty, really?
Example: \(a = 3 \pm 0.5\), where 0.5 is the standard error (jargon: “error” = uncertainty)
What we would like it to mean: “The true value is between 2.5 and 3.5 with a probability of 68 %”
What it technically means (in most cases): “The interval 2.5 to 3.5, if always constructed with the same procedure, contains the true value in 68 % of identically repeated random experiments.”
Both statements coincide only in the asymptotic limit of infinitely large samples
Statistical and systematic uncertainties
Statistical uncertainty
- Origin: We use finite sample instead of infinite distribution
- Goes down as simple size increases
- Example
- Arithmetic mean: \(\bar x = \frac 1N\sum_i x_i\)
- Variance of \(\bar x\): \(\hat V_{\bar x} = \frac{1}{N - 1} \frac 1 N\sum_i (x_i - \bar x)^2\)
- We have reliable standard recipes to calculate these
Variance or standard deviation?
- Standard deviation \(\sigma = \sqrt{V}\)
- Variance is more fundamental
- Second central moment of distribution
- \(V = V_1 + V_2\), but \(\sigma = \sqrt{\sigma_1^2 + \sigma_2^2}\)
- Communicate standard deviations, compute with variances
Systematic uncertainty
- Origin: Imperfect appartus or technique, deviations of reality from simplified models
- Quantifies potential mistakes in analysis
- Does not go down as sample size increases
- Often correlated
- Real-life: Measured the length of ten shoes with ruler than has factory tolerance of 0.1 mm
- LHCb: Measured cross-sections with value of luminosity that has uncertainty of 1-2%
- Not well covered in statistics books, but some theoretical results available
Estimators
Estimator maps dataset to value \(\{x_i \} \to \hat p\)
- Notation: \(\hat p\) is estimate of true value \(p\)
- Estimator can have functional form or be an algorithm
- Arithmetic mean \(\hat \mu = \frac 1N \sum_i x_i\)
- Sample median: Sort \(\{ x_i \}\), pick value in the middle
Many kinds of estimators, some better than others
Optimal estimators
- As close to true value as possible
- Easy to compute and/or easy to apply to any problem
- Robustness against outliers
In HEP, we mainly use
- Plug-in estimator
- Maximum-likelihood estimator (MLE)
Bias and variance
- In one experiment, we obtain sample \(\{ x_i \}\) with size \(N\) and compute estimate \(\hat a\)
- Sample \(\{ x_i \}\) is random, estimate \(\hat a\) also random
- If experiment is identically repeated \(K\) times, we get sample \(\{ x_i \}_k\) and estimate \(\hat a_k\) each time
- Derive accuracy of estimator from estimates \(\{ \hat a_1, \hat a_2, \dots, \hat a_K \}\)
- Bias of \(\hat a\) = \(\frac1K \sum_i (\hat a_i - a)\)
- Variance of \(\hat a\) = \(\frac 1 {K -1}\sum_i (\hat a_i - \frac 1K \sum_k \hat a_k)^2\)
- Example for biased estimator: \(\hat V = \frac1N\sum_i (x_i - \hat\mu)^2\) is biased if \(\hat \mu\) is estimated from same sample
fig, ax = plt.subplots(1, 2, sharex=True, sharey=True, figsize=(10, 4))
ax[0].hist(var_1, bins=50, label="$\hat V_1$");
ax[1].hist(var_2, bins=50, label="$\hat V_2$");
for axi in ax:
axi.axvline(sigma ** 2, color="C3", label="true V")
ax[0].axvline(np.mean(var_1), color="C1", ls="--", label="mean of $\hat V_1$")
ax[1].axvline(np.mean(var_2), color="C1", ls="--", label="mean of $\hat V_2$")
for axi in ax:
axi.legend()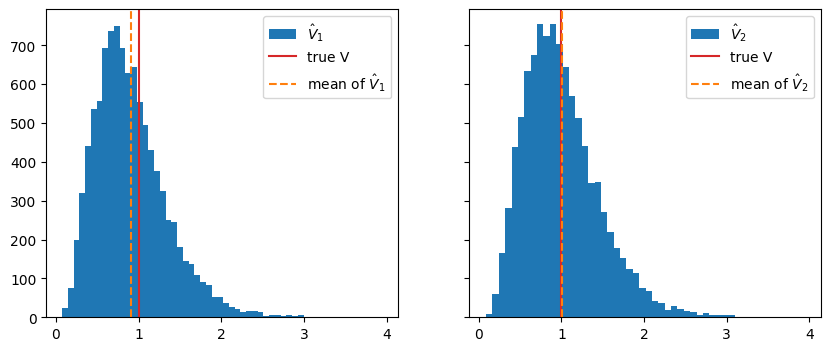
- Well-known bias correction for this case: \[ \hat V_{1,\text{corr}} = \frac N{N-1}\hat V_1 \] For derivation see e.g. F. James book (references at end)
Exercise * Compute \(\hat V_{1, \text{corr}}\) and compare its mean with true \(V\)
# do exercise herePlug-in estimator
- Plug-in principle: replace true values in some formula with estimates, e.g.
\[ c = g(a, b, \dots) \to \hat c = g(\hat a, \hat b, \dots) \]
- Justification
- For \(N\to \infty\), \(\Delta a = \hat a - a \to 0\) for any reasonable estimator
- \(g(\hat a) = g(a + \Delta a) = g(a) + O(\Delta a) \to g(a)\) for smooth \(g\)
- Example: \(\sqrt{N}\) uncertainty estimator for Poisson-distributed count \(N\)
- \(P(N; \lambda) = e^{-\lambda} \lambda^N / N!\)
- Variance of counts: \(V_N = \lambda\)
- Estimator \(\hat \lambda = \frac 1 K \sum_k N_k\); for \(K = 1\), \(\hat \lambda = N\)
- Plug-in principle: \(\hat V_N = \hat \lambda = N\)
- Standard deviation: \(\hat \sigma_N = \sqrt{\hat V_N} = \sqrt{N}\)
- Plug-in estimators can behave poorly in small samples
- Poisson example: \(\lambda = 0.01\), sample \(N = 0 \to \hat \sigma_N = 0\)
Bootstrapping bias and variance of estimators
- Bootstrap method: generic way to compute bias and variance of any estimator
Parametric bootstrap
- Available:
- Sample \(\{ x_i \}\) distributed along \(f(x; a)\), \(a\) is unknown
- Estimator for \(\hat a\)
- Want to know bias and variance of estimator
- Simulate experiment \(K\) times
- Draw samples \(\{ x_i \}_k\) of equal size from \(\hat f(x; \hat a)\) (plugin-estimate)
- Compute \(\hat a_k\)
- Bias \(\frac 1 K \sum_k (\hat a_k - \hat a)\)
- Variance \(\frac 1 {K-1} \sum_k (\hat a_k - \frac 1K \sum_k \hat a_k)^2\)
from numba_stats import norm, truncexpon, poisson
def make_data(s, mu, sigma, b, slope, seed):
sr = poisson.rvs(s, size=1, random_state=seed)[0]
br = poisson.rvs(b, size=1, random_state=seed)[0]
s = norm.rvs(mu, sigma, size=sr, random_state=seed)
b = truncexpon.rvs(0, 1, 0, slope, size=br, random_state=seed)
return np.append(s, b)
truth = {
"s": 1000,
"mu": 0.5,
"sigma": 0.05,
"b": 500,
"slope": 1,
}x = make_data(**truth, seed=0)
w, xe = plt.hist(x, bins=100)[:2];
from iminuit import Minuit
from iminuit.cost import ExtendedBinnedNLL
from numba_stats import norm, truncexpon
def model(x, s, mu, sigma, b, slope):
return s * norm.cdf(x, mu, sigma) + b * truncexpon.cdf(x, 0, 1, 0, slope)
nll = ExtendedBinnedNLL(w, xe, model)
m = Minuit(nll, **truth)
m.limits["mu"] = (0, 1)
m.limits["s", "b", "sigma", "slope"] = (0, None)m.migrad()| Migrad | |
|---|---|
| FCN = 106.9 (χ²/ndof = 1.1) | Nfcn = 101 |
| EDM = 9.95e-07 (Goal: 0.0002) | |
| Valid Minimum | Below EDM threshold (goal x 10) |
| No parameters at limit | Below call limit |
| Hesse ok | Covariance accurate |
| Name | Value | Hesse Error | Minos Error- | Minos Error+ | Limit- | Limit+ | Fixed | |
|---|---|---|---|---|---|---|---|---|
| 0 | s | 1000 | 35 | 0 | ||||
| 1 | mu | 0.4983 | 0.0018 | 0 | 1 | |||
| 2 | sigma | 0.0482 | 0.0014 | 0 | ||||
| 3 | b | 510 | 27 | 0 | ||||
| 4 | slope | 0.95 | 0.15 | 0 |
| s | mu | sigma | b | slope | |
|---|---|---|---|---|---|
| s | 1.2e+03 | -48.4e-6 | 8.9237e-3 (0.177) | -0.2e3 (-0.213) | -0.368 (-0.073) |
| mu | -48.4e-6 | 3.09e-06 | -0 (-0.005) | 61.5e-6 (0.001) | -17.0e-6 (-0.066) |
| sigma | 8.9237e-3 (0.177) | -0 (-0.005) | 2.12e-06 | -8.9641e-3 (-0.231) | -16.5e-6 (-0.078) |
| b | -0.2e3 (-0.213) | 61.5e-6 (0.001) | -8.9641e-3 (-0.231) | 711 | 0.354 (0.091) |
| slope | -0.368 (-0.073) | -17.0e-6 (-0.066) | -16.5e-6 (-0.078) | 0.354 (0.091) | 0.0213 |
- Fit can be written as estimator
def estimator(x):
w = np.histogram(x, bins=xe)[0]
nll = ExtendedBinnedNLL(w, xe, model)
m = Minuit(nll, **truth)
m.limits["mu"] = (0, 1)
m.limits["s", "b", "sigma", "slope"] = (0, None)
m.migrad()
assert m.valid
return m.valuesreplicates = []
for seed in range(100):
x_k = make_data(*m.values, seed=seed+1)
p_k = estimator(x_k)
replicates.append(p_k)
replicates = np.transpose(replicates)Exercise
- Compute bias and standard deviation of replicates for each parameter
- Compare standard deviation of replicates with Minuit’s uncertainty estimate, accessible via
m.errors
# do exercise hereNonparametric bootstrap
- Can do the same by sampling directly from \(\{ x_i \}\)
- Standard method for samples of fixed size \(N\):
- Pick \(N\) random samples from \(\{ x_i \}\) with replacement to obtain \(\{ x_i \}_k\)
- Compute \(\hat a_k\) from \(\{ x_i \}_k\) and proceed as before
- “Extended method” for samples of variable size (Poisson):
- Randomly draw \(M\) from Poisson distribution with \(\lambda = N\)
- Pick \(M\) random samples from \(\{ x_i \}\) with replacement to obtain \(\{ x_i \}_k\)
- …
- Convenient: use same algorithms for any distribution
- Efficient implementations for both algorithms in
resample
import resample
resample.__version__'1.7.1'from resample.bootstrap import resample
# generate replicated samples of varying size (Poisson distributed)
for x_k in resample(x, size=100, method="extended", random_state=1):
print(x_k)
break[0.58820262 0.58820262 0.52000786 ... 0.38532216 0.26892968 0.26892968]Exercise
- Again compute replicated fit results using
resampeand store results in a list - Copy previous code where possible
- Again compute bias and standard deviation of replicates and compare with Minuit’s uncertainty estimate
# do exercise hereresampleprovides shortcut for computing variance
from resample.bootstrap import variance
var = variance(estimator, x, method="extended", size=100, random_state=1)
for i, name in enumerate(m.parameters):
std = var[i] ** 0.5
print(f"{name:8} 𝜎(Minuit) = {m.errors[i]:6.2g} 𝜎(bootstrap) = {std:6.2g}")s 𝜎(Minuit) = 35 𝜎(bootstrap) = 33
mu 𝜎(Minuit) = 0.0018 𝜎(bootstrap) = 0.0017
sigma 𝜎(Minuit) = 0.0015 𝜎(bootstrap) = 0.0014
b 𝜎(Minuit) = 27 𝜎(bootstrap) = 27
slope 𝜎(Minuit) = 0.15 𝜎(bootstrap) = 0.17Exercise
- Repeat the variance calculation without using
method="extended" - Compare the uncertainties of \(s\) and \(b\)
# do exercise here- Bootstrap can be applied to most statistical problems in HEP
- Restriction: samples must be identically and independently distributed
- May work well even with small samples (< 10)
Error propagation
- Full analysis typically combines many intermediate results with uncertainties into final result
- Consider \(\vec b = g(\vec a)\) with uncertainties in \(\vec a\)
- Two ways of computing uncertainties of \(\vec b\)
- Monte-Carlo simulation
- Error propagation
- Can be equally used for statistical and systematic uncertainties
Monte-Carlo simulation
Draw variations of fit result from multivariate normal distribution
Compute variable of interest from variations
Compute standard deviation or covariance of
Example: compute uncertainty of \(\hat N = \hat s + \hat b\)
rng = np.random.default_rng(1)
a_k = rng.multivariate_normal(m.values, m.covariance, size=1000)
def g(a):
return a[0] + a[3]
b_k = np.apply_along_axis(g, -1, a_k)
print(f"𝜎(N, simulation) = {np.std(b_k):.3g}")
print(f"𝜎(N, naive) = {(m.errors[0]**2 + m.errors[3]**2)**0.5:.3g}")
print(f"𝜎(N, expected) = {np.sqrt(len(x)):.3g}")𝜎(N, simulation) = 37.7
𝜎(N, naive) = 43.7
𝜎(N, expected) = 38.8- \(\sigma_{N,\text{simulation}}^2 < \sigma_s^2 + \sigma_b^2\), because \(s_k\) and \(b_k\) are anti-correlated
Error propagation
Given: \(\vec a\) with covariance matrix \(C_a\) and differentiable function \(\vec b = g(\vec a)\)
\(\vec b\) can have different dimensionality from \(\vec a\)
One can derive via first-order Taylor expansion: \[ C_b = J \, C_a \, J^T \text{ with } J_{ik} = \frac{\partial g_i}{\partial a_k} \]
Useful special cases
- Sum of scaled independent random values \(c = \alpha a + \beta b\) (\(\alpha, \beta\) are constant) \[ \sigma^2_c = \alpha^2 \sigma^2_a + \beta^2 \sigma^2_b \]
- Product of powers of independent random values \(c = a^\alpha \, b^\beta\) \[ \left(\frac{\sigma_c}{c}\right)^2 = \alpha^2 \left(\frac{\sigma_a}{a}\right)^2 + \beta^2 \left(\frac{\sigma_b}{b}\right)^2 \]
Final uncertainty often dominated by one term
When estimating systematic uncertainties: put your effort in estimating largest term and be lax on others
Computation of \(J\) by hand error-prone and tedious
jacobilibrary offers automatic error propagation based on a robust numerical derivative

import jacobi
jacobi.__version__'0.9.2'b, cov_b = jacobi.propagate(g, m.values, m.covariance)
f"𝜎(N, error propagation) = {cov_b ** 0.5:.3g} 𝜎(N, expected) = {np.sqrt(len(x)):.3g}"'𝜎(N, error propagation) = 38.9 𝜎(N, expected) = 38.8'jacobi.propagateworks for any differentiable mapping, which even includes most fits!
from iminuit.cost import LeastSquares
def h(pvec):
a, b = pvec
x = np.array([1, 2, 3])
y = a + b * x
m = Minuit(LeastSquares(x, y, 1, lambda x, a, b: a + b * x), a=0, b=0)
m.strategy = 0
m.migrad()
assert m.valid
return m.values
jacobi.propagate(h, (1, 2), (1, 1))(array([1., 2.]),
array([[ 1.00000000e+00, -1.84267416e-12],
[-1.84267416e-12, 1.00000000e+00]]))jacobi.propagatemakes complex error propagation feasible- Extensively used in the pp @ 13 TeV analysis of prompt charged particle production
Systematic uncertainties: Known unknowns
“There are known knowns, things we know that we know;
and there are known unknowns, things that we know we don’t know.
But there are also unknown unknowns, things we do not know we don’t know.”
Donald Rumsfeld
- Why data analyses take so long: study data, detector, and methods to turn unknown unknowns into known knowns or known unknowns
- known unknowns = systematic uncertainties
- No formal way, but generally…
Worry * What could go wrong? * What are my assumptions?
Example * You take an efficiency or correction factor from simulation, but simulation differs from real experiment. * Solution: Perform control measurement to estimate how much simulation differs from real experiment.
Methods * Use methods that are known to be optimal for your problem or derive them from first principles * General purpose methods often safer to use than special-case methods * Maximum-likelihood fit > least-squares fit * Bootstrap estimate of uncertainty > other uncertainty estimates
Apply checks * Validate whole analysis on simulated data (sometimes you can use toy simulation) * Split data by magnet polarity, fill, etc., analyze splits separately, check for agreement
Guidelines for handling systematic errors * Roger Barlow: Systematic Errors: Facts and Fictions (2002) * Roger Barlow, “Practical Statistics for Particle Physics” (2019), Section 6.5
Further resources for learning
LHCb Statistics and ML WG
Statistics Guidelines to collect information about best practices
Introductory statistics books
List of books on TWiki pages of Statistis and Machine Learning WG
Books I personally like a lot
- Glen Cowan, “Statistical Data Analysis”, Oxford University Press, 1998
- F. James, “Statistical Methods in Experimental Physics”, 2nd edition, World Scientific, 2006
- R. Barlow, “Statistics: a Guide to the Use of Statistical Methods in the Physical Sciences”, Wiley, 1989
- B. Efron, R.J. Tibshirani, “An Introduction to the Bootstrap”, CRC Press, 1994
Lectures, papers, notebooks
- Roger Barlow
- More Jupyter notebooks
- All about sWeights and COWs with gentle derivation
- Machine learning