#!pip install scipy matplotlib numpy scikit-learn xgboostHans Dembinski
Overview

Caution: Field is riddled with buzzwords
- Machine learning, artificial intelligence, deep learning, belief networks, attention-based networks, long short-term memory, features, hyperparameters …
Anthropomorphized and technical names hide what are usually basic mathematical algorithms
Machine learning: umbrella term for multivariate statistical methods for classification and regression
- Multivariate: algorithms use more than one variable as input, e.g. (x, y, z)
- Statistical: algorithms can handle noisy input and are founded on statistical theory
- Classification: sort inputs based on input variables into categories
- Regression: find a mapping of input vectors to output vectors (fitting)

- Rise of the machines: Why do we need them?
- Humans good at working with 1D or 2D data, because we can visualize it
- Humans bad at working with higer-dimensional data
- Algorithms (“machines”) scale well to high-dimensional data
Notable software
- Multiple algorithms and utilities
- Gradient-boosted decision trees (aka gradient boosting machines)
- Neural networks
- Tensorflow
- Keras (high-level interface to Tensorflow)
- PyTorch
- …
Learning resources
Internet provides excellent resources for autodidactic learning
- Example code
- Tutorials
- Documentation
All methods can do classification and regression and work with any input
What to use?
- Structured data (i.e. tables of numbers): Ensembles of decision trees
- Good results with default settings
- Can process continuous and categorical input (m/w, red/green/blue, …)
- No data preprocessing required
- Compute feature importance
- Unstructured data (i.e. images, waveforms, text…): Neural networks
- Deep networks learn to extract features automatically
- Requires data preprocessing
- Networks difficult to optimize for non-experts
- Often need very large data sets for training
- Structured data (i.e. tables of numbers): Ensembles of decision trees
Some definitions * sample: one or more numbers which describe outcome of stochastic process * dataset: consists of samples * features: subset of numbers from each sample which carry interesting information, also numbers computed from each sample * signal: samples which originate from stochastic process of interest, typically particle decay * background: samples which do not originate from process of interest
Discuss how the typical decay \(K^0_S \to \pi^+ \pi^-\) is reconstructed and why in general there is always some background.
Clarify how the previous terms would be used in the context of this example.
Classification
- Setting: dataset is mix of two or more components, typically signal + one or more backgrounds
- Want only signal component, invert a cut to select signal candidates
- Features are variables which allow to discriminate between signal and background, for example, reconstructed mass of decay candidate
import scipy
import matplotlib
import numpy
import sklearn
import xgboost
print(f"scipy {scipy.__version__}")
print(f"matplotlib {matplotlib.__version__}")
print(f"numpy {numpy.__version__}")
print(f"sklearn {sklearn.__version__}")
print(f"xgboost {xgboost.__version__}")scipy 1.10.0
matplotlib 3.7.1
numpy 1.21.6
sklearn 1.2.2
xgboost 1.7.4from scipy.stats import norm
from matplotlib import pyplot as plt
import numpy as np
background = norm(0, 1).rvs(1000, random_state=1)
signal = norm(1, 0.1).rvs(500, random_state=1)
x = np.append(signal, background)
y = np.append(np.ones(len(signal)), np.zeros(len(background)))def plot(x, y):
signal = x[y == 1]
background = x[y == 0]
t, xe = np.histogram(x, bins=100)
s = np.histogram(signal, bins=xe)[0]
b = np.histogram(background, bins=xe)[0]
plt.stairs(b, xe, fill=True, label="background")
plt.stairs(t, xe, baseline=b, fill=True, label="signal")
plt.xlabel("x")
plt.legend();
return xe
plot(x, y);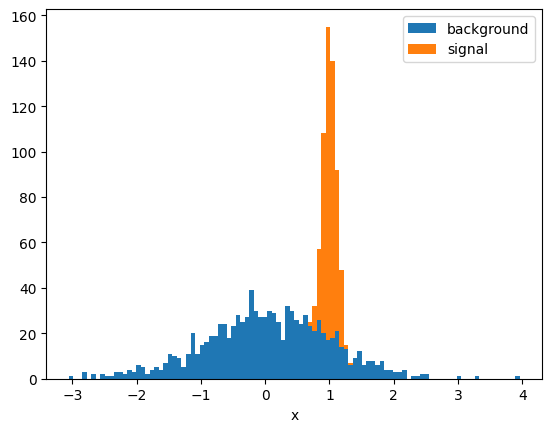
Exercise: * Propose simple selection (“cut”) that keeps most of the signal and removes most of the background * Is it possible to eliminate the background completely?
Efficiency and purity of a selection
- Efficiency: fraction of signal left after applying selection
- Number of true signal samples after cut / Number of all true signal samples
- Purity: fraction of background still contained in selected sample
- Number of true signal samples after cut / Number of selected samples
- Want to maximize efficiency and purity
Single-parameter cut
xmin = 0.7
selected_signal = np.sum(x[y==1] > xmin)
total_signal = np.sum(y==1)
selected = np.sum(x > xmin)
efficiency = selected_signal / total_signal
purity = selected_signal / selected
plot(x, y)
plt.axvspan(-3.5, xmin, facecolor="k", alpha=0.5)
plt.legend(title=f"efficiency={efficiency:.2f}\npurity={purity:.2f}");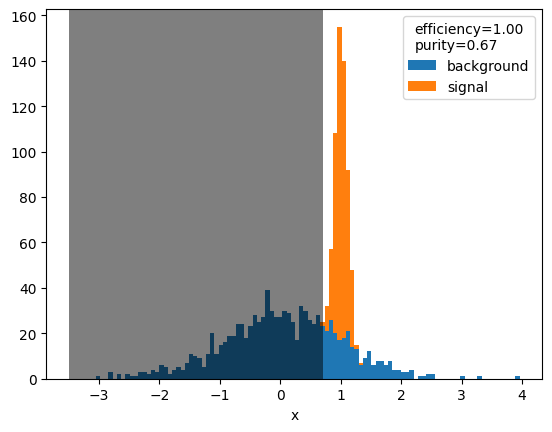
xmin = np.linspace(-3, 3, 100)
efficiency = []
purity = []
for xm in xmin:
selected_signal = np.sum(signal > xm)
total_signal = len(signal)
selected = np.sum(x > xm)
e = selected_signal / total_signal
p = selected_signal / selected
efficiency.append(e)
purity.append(p)
plt.plot(xmin, efficiency, label="efficiency")
plt.plot(xmin, purity, label="purity")
plt.xlabel("xmin")
plt.legend();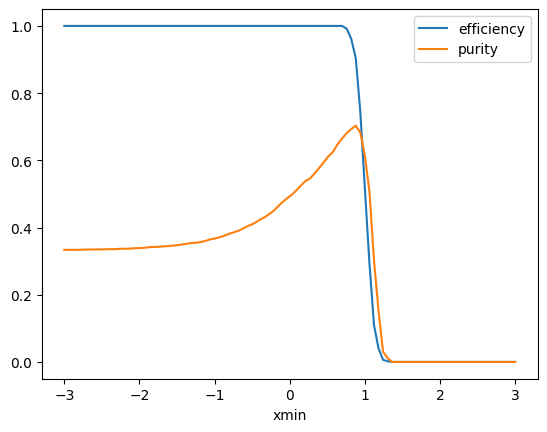
plt.scatter(efficiency, purity, c=xmin);
plt.xlim(0, 1)
plt.ylim(0, 1)
plt.xlabel("efficiency")
plt.ylabel("purity")
plt.colorbar().set_label("xmin")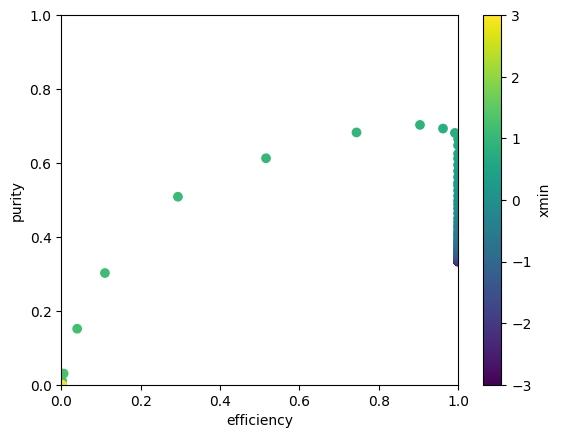
arg = np.argmax(purity)
print("efficiency at max purity", efficiency[arg])efficiency at max purity 0.904How to choose cut?
- Want to maximize efficiency and purity, but what trade-off between the two?
Often subjective choice is acceptable, because exact answer is difficult
Objective answer given by figure-of-merit (FOM)
- Popular is Punzi FOM
- Most FOM (including Punzi FOM) only minimize statistical uncertainty
- Correct FOM has to minimize total uncertainty = statistical ⊕ systematic
- Systematic uncertainty often difficult to quantify: fallback to Punzi or subjective choice
BDT-based cut
We can do better with two cuts, on xmin and xmax
Space of cut parameters then already two-dimensional
With 2D data
- Four rectangular cuts possible (4 cut parameters)
- Infinitely many non-rectangular cuts possible
Use machine instead, needs to be trained on labeled data
Use BDT classifier from XGBoost
Gradient boosting machines regularly win Kaggle competitions
Well documented, fast, parallizable, tunable
from xgboost.sklearn import XGBClassifier
from sklearn.model_selection import train_test_split
# generate training data, ideally we have more than actual data
background = norm(0, 1).rvs(100000, random_state=2)
signal = norm(1, 0.1).rvs(50000, random_state=2)
x2 = np.append(signal, background)
# features in general matrix [N, K] for N samples and K features
X = x2.reshape((len(x2), 1))
# labels: signal=1, background=0
y = np.append(np.ones_like(signal), np.zeros_like(background))# ALWAYS do this: split into train and test datasets
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=3)
# actual magic happens here
clf = XGBClassifier(random_state=4)
clf.fit(X_train, y_train);fig, ax = plt.subplots(2, sharex=True)
plt.sca(ax[0])
xe = plot(X_test[:, 0], y_test)
plt.xlabel(None) # unset label set by `plot`
plt.sca(ax[1])
score = clf.score(X_test, y_test)
xm = np.linspace(xe[0], xe[-1], 200)
ps, pb = clf.predict_proba(xm).T
plt.xlabel("x")
plt.plot(xm, ps, "C0-", label="background probability")
plt.plot(xm, pb, "C1-", label="signal probability")
plt.legend(title=f"score={score:.2f}", frameon=False);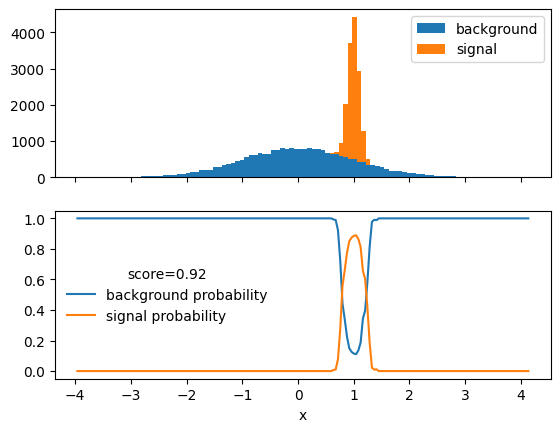
Exercise: * Vary some hyperparameters in XGBClassifier to see how score, runtime, and predicted probability changes * n_estimators try e.g. 2, 5, 10 * max_depth try e.g. 1, 5, 10 * learning_rate try e.g. 0.001, 0.05, 0.5, 1
- BDT predicts signal probability
- We use this to implement our cut: select regions with high signal probability
ps = clf.predict_proba(X_test[y_test==1])[:, 1]
pb = clf.predict_proba(X_test[y_test==0])[:, 1]
xe = plt.hist(ps, bins=20, alpha=0.5, label="true signal")[1]
plt.hist(pb, bins=xe, alpha=0.5, label="true background")
plt.legend()
plt.xlabel("predicted signal probability")
plt.ylim(0, 5e3);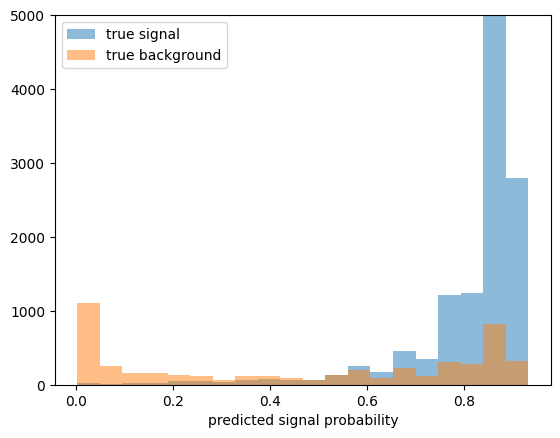
xe = plot(X_test[:, 0], y_test)
# chosen cut: signal probability > 0.6
# visualize accepted region
xm = np.linspace(xe[0], xe[-1], 200)
mask = clf.predict_proba(xm)[:, 1] > 0.6
plt.plot(xm, mask * 5000, "k-");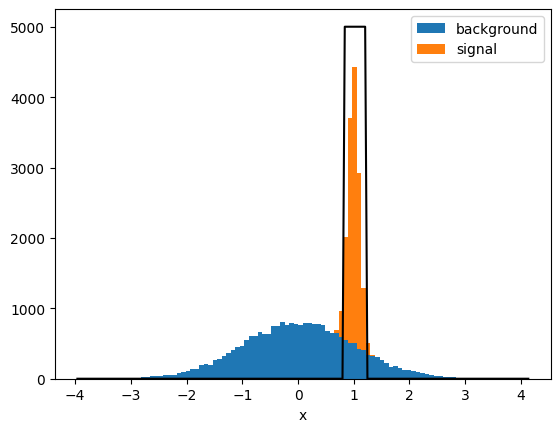
pmin = np.linspace(0, 1, 100)
efficiency = []
purity = []
for pm in pmin:
mask = clf.predict_proba(X_test)[:, 1] > pm
selected = np.sum(mask)
selected_signal = np.sum(y_test[mask])
total_signal = np.sum(y_test)
efficiency.append(selected_signal / total_signal)
with np.errstate(divide="ignore", invalid="ignore"):
purity.append(selected_signal / selected)
plt.plot(pmin, efficiency, label="efficiency")
plt.plot(pmin, purity, label="purity")
plt.xlabel("pmin")
plt.legend();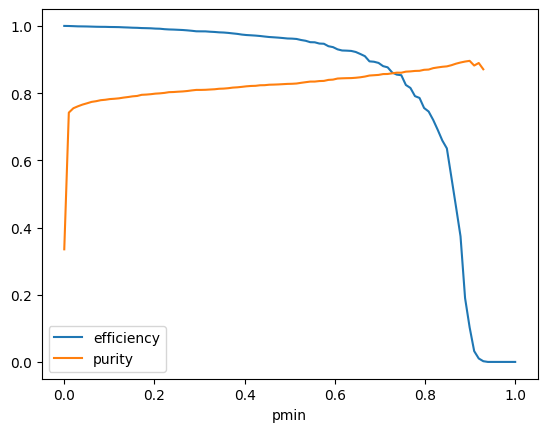
plt.scatter(efficiency, purity, c=pmin);
plt.xlim(0, 1)
plt.ylim(0, 1)
plt.xlabel("efficiency")
plt.ylabel("purity")
plt.colorbar().set_label("pmin")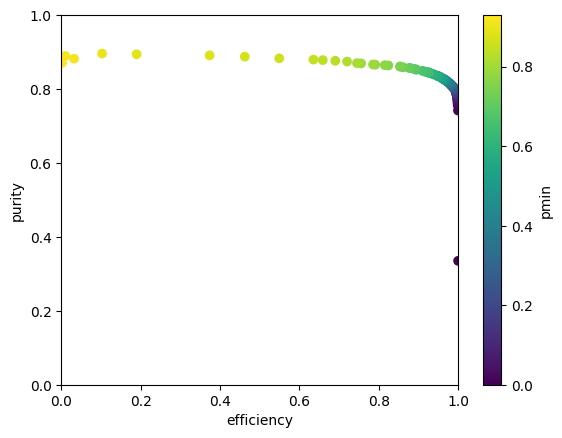
- Physicists like efficiency and purity, but ROC curve is standard
- Receiver-operator characteristic
- ROC plots true positive rate (fraction of signal that passes cut) over false positive rate (fraction of background that passes cut)
- Scikit-Learn provides a quick way to generate this plot
from sklearn.metrics import RocCurveDisplay
RocCurveDisplay.from_estimator(clf, X_test, y_test);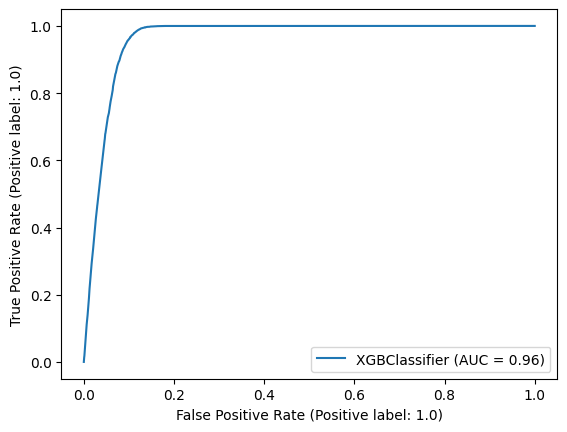
- area under curve (AUC) is a common metric to compare the performance of two different classifiers
- can also use Scikit-Learn to compute ROC points and AUC and plot these ourselves
from sklearn.metrics import roc_curve, auc
# plot XGBoost curve again
fpr, tpr, thresholds = roc_curve(y_test, clf.predict_proba(X_test)[:, 1])
plt.plot(fpr, tpr, label=f"XGBoost AUC={auc(fpr, tpr):.2f}")
# plot ROC curve for our simple x > 0.7 cut for comparison
fpr2, tpr2, _ = roc_curve(y_test, X_test[:, 0] > 0.7)
plt.plot(fpr2, tpr2, label=f"x > 0.7 AUC={auc(fpr2, tpr2):.2f}")
plt.xlabel("False positive rate")
plt.ylabel("True positive rate")
plt.legend();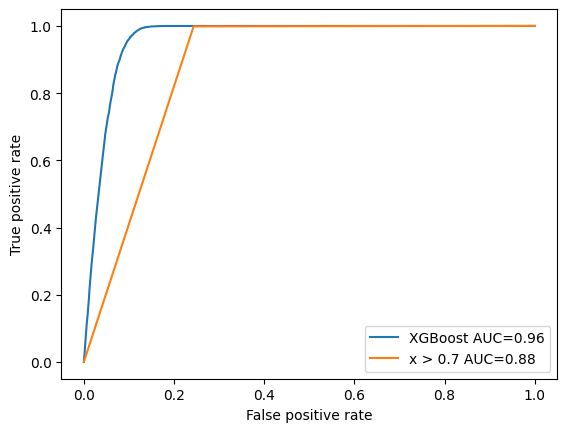
- Exercise: run scikit-learn example
- Try adding XGBClassifier into the comparison
- Play around with the hyperparameters of the classifiers
Regression
Regression (aka fitting): “learn” function y = f(x) from points(x, y), where both x and y can be vectors
Machines can do this, but we rarely need this in HEP
We usually prefer to use theoretically motivated models or hand-crafted low-order polynoms
Let’s try regression on one-dimensional function
def model(x, a, b, c, d):
return a + b * np.sin(c * x) * np.exp(- d * x)
truth = 0.2, 1, 2, 0.5
x = np.linspace(0, 10, 1000)
plt.plot(x, model(x, *truth))
plt.xlabel("x")
plt.ylabel("y");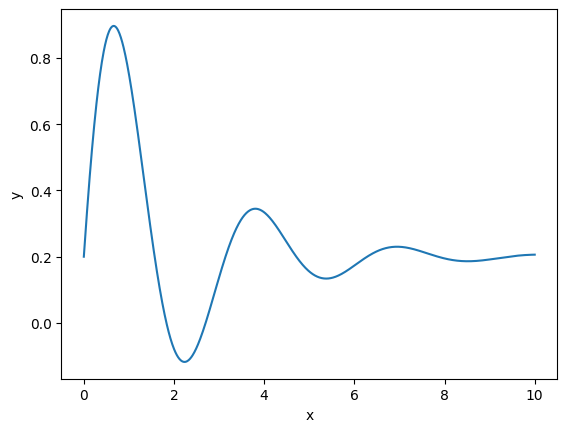
- generate random points on this curve and add some noise to make it more realistic
rng = np.random.default_rng(1)
x = rng.uniform(0, 10, 1000)
y = model(x, 0.2, 1, 2, 0.5)
y += rng.normal(0, 0.05, len(y))
X = np.reshape(x, (-1, 1))- (X, y) are inputs for regression similar to before, but y is a real number now, not a label (integer)
Neural network
- We use MLPRegressor from Scikit-Learn, a basic neural network
- Neural networks always need preprocessing
- Scikit-Learn offers pipelines to automate this, use them
from sklearn.neural_network import MLPRegressor
from sklearn.preprocessing import StandardScaler
from sklearn.pipeline import make_pipeline
# network with 5 hidden layers with 100 nodes each
regressor = MLPRegressor(
hidden_layer_sizes=(100,)*5,
random_state=1)
# we use basic StandardScaler for preprocessing
pipeline = make_pipeline(StandardScaler(), regressor)
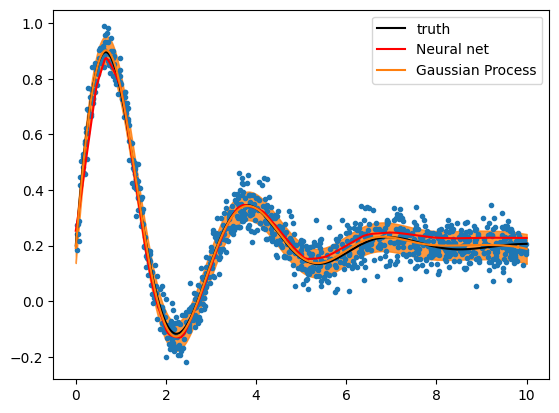
pipeline.fit(X, y);# compute fitted curve
X2 = np.reshape(np.linspace(0, 10, 1000), (-1, 1))
y2 = pipeline.predict(X2)
plt.plot(x, y, ".")
plt.plot(X2[:, 0], y2, color="r", label="Neural net")
plt.plot(X2[:, 0], model(X2[:, 0], *truth), color="k", label="truth")
plt.legend()
plt.xlabel("x")
plt.ylabel("y");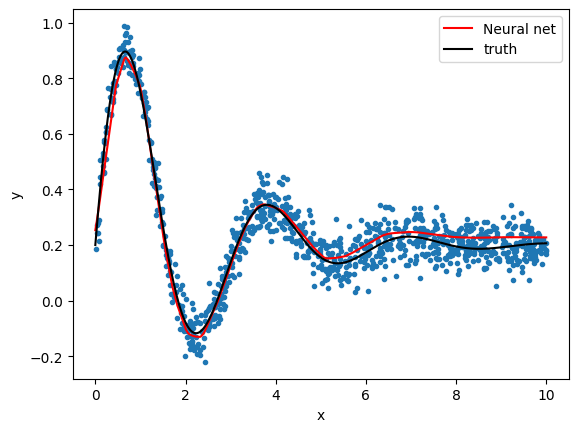
- Exercise: play around with hyperparameters of MLPRegressor
- What happens if you remove the StandardScaler?
- What performs better, deep or wide networks?
Gaussian Process
- For low-dimensional data, Gaussian Process Regression can work well
- Not easy to set up: need to select suitable kernels and configure them
- Advantage: Predicts uncertainty and automatically provides smooth solution
from sklearn.gaussian_process import GaussianProcessRegressor
from sklearn.gaussian_process.kernels import RBF, WhiteKernel
kernel = RBF() + WhiteKernel()
gpr = GaussianProcessRegressor(kernel=kernel)
gpr.fit(X, y)
y3, y3err = gpr.predict(X2, return_std=True)plt.plot(x, y, ".")
plt.plot(X2[:, 0], model(X2[:, 0], *truth), color="k", label="truth")
plt.plot(X2[:, 0], y2, color="r", label="Neural net")
l = plt.plot(X2[:, 0], y3, label="Gaussian Process")[0]
plt.fill_between(X2[:, 0], y3 - y3err, y3 + y3err, alpha=0.8, color=l.get_color())
plt.legend();