from resample.jackknife import jackknife
import numpy as np
import numba as nb
from typing import Callable
from timeit import timeit
from matplotlib import pyplot as pltMy resample library implements the jackknife and bootstrap resampling techniques. The implementations use pure-Python. Since resampling techniques are computationally expensive, it is important to ask whether we can speed up these methods with Numba. In this notebook I compare a JIT-compiled implementation of the Jackknife with the pure Python implementation from resample.
Since Numba currently does not support JIT-compiled functions which accept other functions as arguments, the JIT-compiled version has to work a little differently from the pure-Python version. It creates a two-dimensional array with all leave-one-out samples at once inside the compiled code. This array is then passed to Python and the function is evaluated over this array. This has a downside: For large samples, creating an N x (N-1) array in memory becomes dangerous since the memory allocation may bring the computer down and very slow.
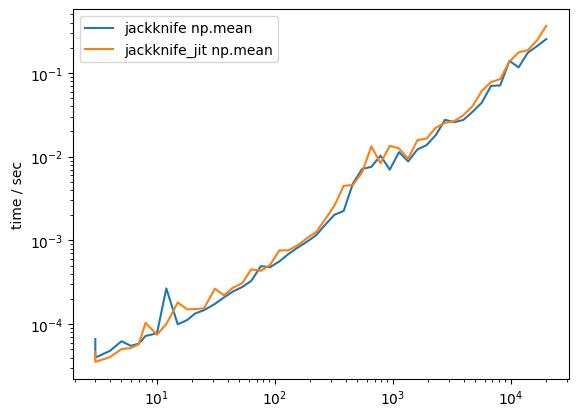
Fortunately, it turns out that there is nothing to gain from JIT-compiling and thus there is no need to accept this caveat.
@nb.njit(nogil=True)
def _jackknife_jit(a: np.ndarray):
n = len(a)
x = np.empty((n, n - 1), dtype=a.dtype)
for i in range(n):
for j in range(n-1):
shift = 0 if j < i else 1
x[i, j] = a[j + shift]
return x
@nb.njit
def _jackknife_gen(a):
n = len(a)
x = np.empty(n - 1, a.dtype)
for i in range(n - 1):
x[i] = a[i + 1]
yield x.view(x.dtype)
# update needs to change values only up to i
# for a = [0, 1, 2, 3]
# x0 = [1, 2, 3]
# x1 = [0, 2, 3]
# x2 = [0, 1, 3]
# x3 = [0, 1, 2]
for i in range(1, n):
for j in range(i):
x[j] = a[j]
yield x.view(x.dtype)
def jackknife_jit(fn: Callable, a: np.ndarray) -> np.ndarray:
return np.asarray([fn(x) for x in _jackknife_gen(a)])
# force compilation
jackknife_jit(np.mean, np.random.randn(10));x = np.random.rand(10000)%%timeit
jackknife(np.mean, x)105 ms ± 10.6 ms per loop (mean ± std. dev. of 7 runs, 10 loops each)%%timeit
jackknife_jit( np.mean, x)119 ms ± 6.34 ms per loop (mean ± std. dev. of 7 runs, 10 loops each)The results are comparable. This also remains so when we benchmark samples of varying size.
n = np.asarray(np.geomspace(3, 20000, 50), dtype=int)
times = {
("jackknife", "np.mean"): np.empty(len(n)),
("jackknife_jit", "np.mean"): np.empty(len(n)),
}
for i, ni in enumerate(n):
x = np.random.randn(ni)
for s_fcn, s_arg in times:
t = timeit(f"{s_fcn}({s_arg}, x)", f"from __main__ import {s_fcn}, x; import numpy as np", number=1)
times[(s_fcn, s_arg)][i] = tplt.figure()
for (s_fcn, s_arg), ts in times.items():
plt.plot(n, ts, label=f"{s_fcn} {s_arg}")
plt.loglog()
plt.ylabel("time / sec")
plt.legend();